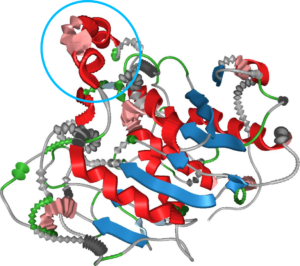
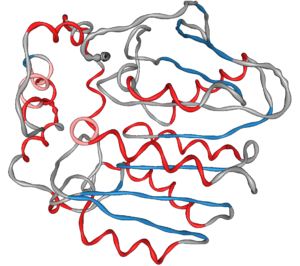
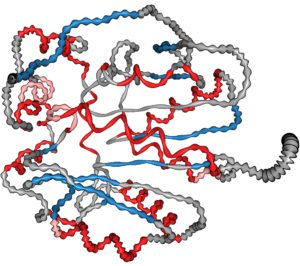
We presented an approach for uncertainty visualization of proteins at the 11th IEEE Pacific Visualization Symposium in Kōbe, Japan.
In some application areas such as medical visualization or weather forecasting, uncertainty is nowadays often illustrated to enable a more informed visual analysis. However, molecular visualization rarely incorporates uncertainty, although it is present in the data.
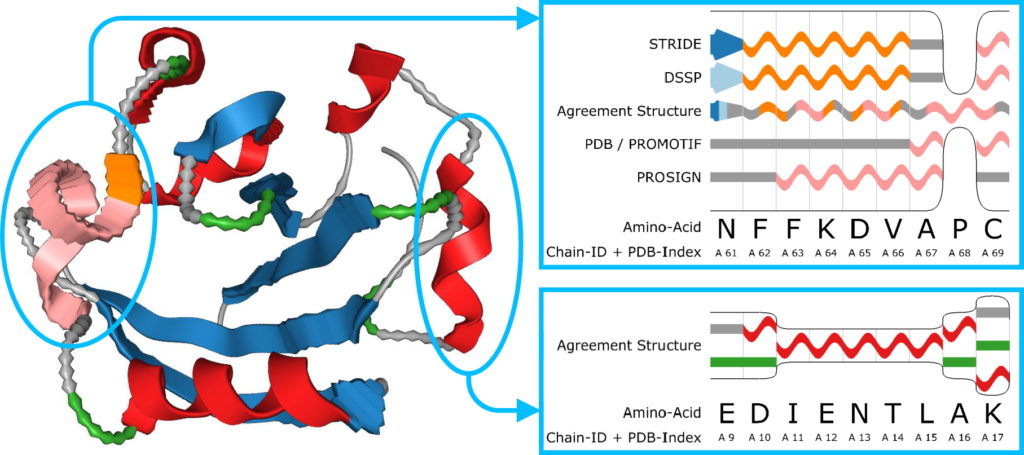
Therefore, our goal was to develop a technique that conveys the uncertainty in the secondary structure of proteins without interfering with de-facto standards present in structural biology. This means that neither the spatial location of the protein chain nor the color scheme may be used to convey uncertainty.
Hence, we use geometric distortion to convey various types of uncertainty such as diverging classification schemes, aggregation over time and flexibility.
Reference:
C. Schulz, K. Schatz, M. Krone, M. Braun, T. Ertl and D. Weiskopf, Uncertainty Visualization for Secondary Structures of Proteins, 2018 IEEE Pacific Visualization Symposium (PacificVis), Kobe, 2018, pp. 96-105.